For 0.11.0
Previous: Working With PHAT Projects

Read Files
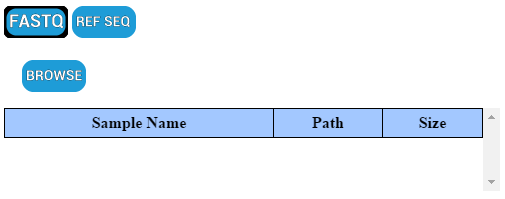
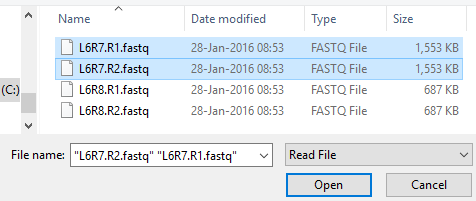
PHAT’s purpose is to make it easier to work with read files (.fastq) and reference sequences (.fasta, .fa, etc); as such all aspects of PHAT revolve around these files. To input read files into PHAT, click “INPUT” on the toolbar. In the input window, click “BROWSE”. By default, the file picker will only show you read files.

Reference Sequences
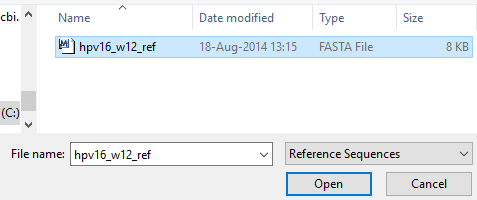
Inputting reference sequences follows the same process. Click the “Ref Seq” button to change the input window to the Reference Sequence view.
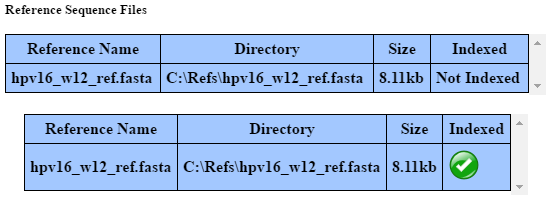
Before reference sequences can be used for alignment, they must be indexed. Clicking on the text “Not Indexed” on an unindexed reference sequence will begin to index it. This process can take some time depending on the size of the file as well as the power of your computer. A good rule is to have some amount of RAM greater than or equal to the size of the file you want to index. If you are trying to index large files (greater than 500MB), it is recommended to close all other applications on your computer while the operation runs.
Before reference sequences can be used for visualization (including being linked to alignment maps inputted externally), they must be indexed for visualization. This operation is generally very fast and separate from indexing for alignment.
Alignment Maps
PHAT can visualize and analyze alignment maps (.sam or .bam) produced on other platforms such as Galaxy. To import an alignment map, click the “Align” button to change the input window to the Alignment view. Before alignment maps can be visualized or analyzed, they must be “linked” to a reference sequence. Click on the “Ref Seq” column for an alignment map you want to link. A list of potientially compatible and potentially incompatible reference sequences will be presented. Click on the “Link This Ref” column for the reference you want to link to. If the reference you want to link to is listed under “Incompatible References”, click on the “Reason” column to find out why. Note: PHAT only compares contig names between references and alignment maps to determine compatibility and its predictions may therefore not always be accurate. Let us know how to make it better.
After an alignment map has been inputted and linked to a reference sequence, it will appear in the Output window as well as the Genome Builder as a visualization option for the reference it was linked to.
Headerless Sequence Alignment Maps
Inputting a .sam file without a header will fail. If inputting has failed, a prompt will appear where you can select a reference sequence to use to substitute header information. If you have not input any references yet, leave the prompt open and input them. They will appear in the prompt after being input. They do not need to be indexed first. If you select a reference which was not used to build the alignment map then the result of inputting the alignment will not be useable. At no point is the alignment map being input actually modified. You can input it again with the correct reference. Note: Be careful with bulk importing many .sam files at once. A prompt will appear for every .sam lacking header information.
Working With Inputs
PHAT makes a distinction between “selected” and “unselected” inputs. By default, files that are input into PHAT are “selected”. Those inputs which are “selected”, will appear for use across PHAT. Those things which are results of multiple inputs (such as alignments), require all of their inputs to be “selected” in order for their results to appear. Those inputs which are “unselected” will not appear in other parts of PHAT. A given input can be identified as “selected” in the input window by appearing bolded in its respective table, and unbolded if “unselected”.

PHAT makes a distinction between inputs which are “imported” and “not imported”. By default, when you input a file it is “not imported”. Inputs can be identified as “not imported” by having their absolute path under the “Path” column in its respective table.
For those read and reference files which are “not imported”, if they are moved, deleted or otherwise modified since they were input into PHAT, trying to perform other operations with them may break or have unexpected results. The exception to this rule for reference sequences is sequence visualization either through PHAT’s circular genome builder, or alignment viewing through the pileup viewer.
If a project has files which are “not imported”, and the project is exported, the same stipulations above will apply for those files which are “not imported”.
When a file is “imported”, it is physically copied into the project and the above stipulations no longer apply. To “import” all the inputs which are selected, scroll to the bottom of the input window and click “IMPORT”. Inputs can be identified as “imported” by the the text “In Project” under the “Path” column in their respective table.
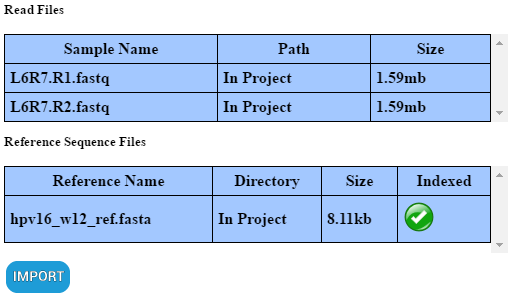
Before you export a project, ensure that those inputs which you want those you share the project with to be to able to run analyses are “imported”. Any inputs which where “not imported” into the project prior to exporting will not be present and those you share the project with will only be able to view past analyses.
Next: Running Quality Control (QC) Reports
About Inputs
When a file is input into PHAT, PHAT maintains a record of its location in the filesystem and little else. For operations which work with files directly (such as QC reports for read files, alignments, etc), PHAT passes the operation in question the absolute path to the files required as they existed when they were first input. Results from operations are stored within the project itself. This allows a project’s results to be shared without having to worry about lugging around potentially large primary datasets. File importing is handled as a special case. In addition to the results of a reference sequence being indexed, information about each contig in that sequence is stored in the project as well. This what allows them to be visualized even if they have been moved or not imported before exporting the project. Alignment maps are copied into the project when input and are treated as though they were an alignment run from within PHAT.
See: file module, file import operation.