For 0.11.0
Previous: Circular Visualization



BLASTing Alignments
PHAT provides automated submission, retrieval and interpretation of NCBI BLAST reports for the purposes of genotyping. PHAT will look specifically for unaligned fragments embedded inside of larger, aligned reads in alignments run from within PHAT as well as imported SAM/BAM files. Those identified reads and their fragments are BLASTed.
Selecting a Reference Sequence Range to BLAST
Typically, only particular regions of an alignment are of interest. PHAT BLASTs operate on sub-sequences. Sequence selection for a given reference genome is performed through the Genome Builder.

Selecting “Figure Actions”, “Select Sequence” on an open figure will began an interactive sequence selection interface.
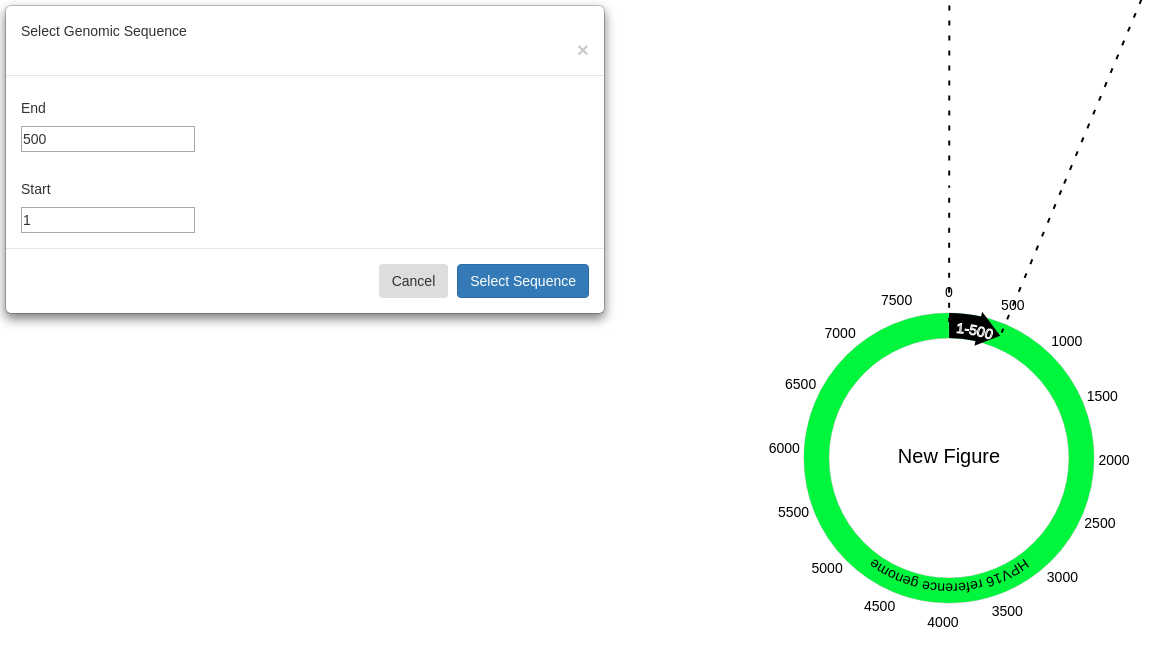 By default, the selected sequence will be from 1-500 on the reference genome. Adding coverage tracks for the alignments(s) of interest will help to narrow down the area of interest though is not required.
By default, the selected sequence will be from 1-500 on the reference genome. Adding coverage tracks for the alignments(s) of interest will help to narrow down the area of interest though is not required.
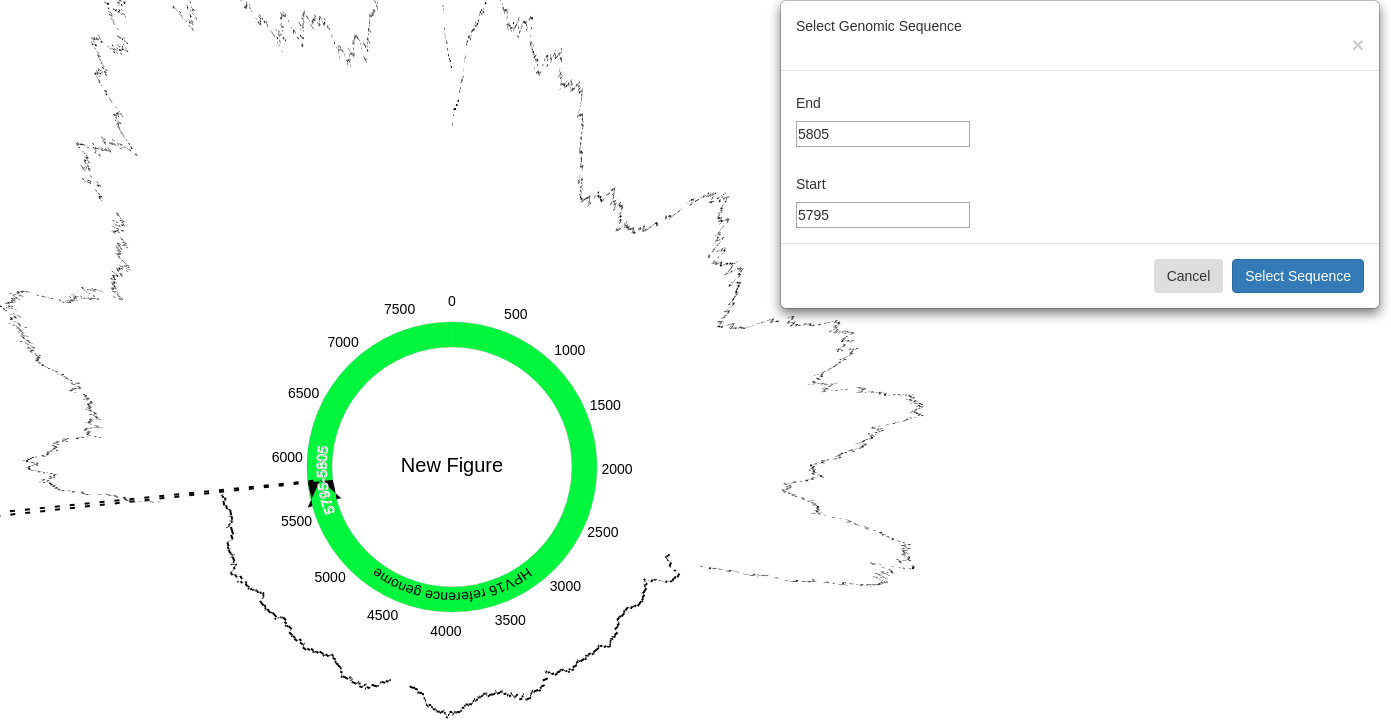
Once a sequence has been selected, clicking “BLAST an Alignment from Start to End” (where Start and End are the selected sequence’s start and end points, respectively) will allow you to then select an alignment to BLAST. Select “BLAST” under the “Options” column of the desired alignment.
Search Algorithm
For the selected alignment, reads which aligned starting between the selected start and end points, and which are 30nt or greater and contain one or more fragments which did not align, of which one fragment must be 30nt or greater (a “suspicious” read) are BLASTed against the nt database. Each unaligned fragment in every “suspicious” read which is 30nt or greater (a “suspicious” fragment) is BLASTed against the nt database. If no hits are found for a given “suspicious” fragment in nt, the fragment is “re-BLASTed” against the Human G+T database. Only the topmost hit is reported for the read and its fragment(s) regardless of database.
Viewing BLAST Results
PHAT will report progress on a particular “BLAST Run” (the algorithm described above) in terms of identified “suspicious” reads and BLASTed “suspicious” fragments and reads. When complete, results can be viewed by clicking on the “BLAST Runs” column for the BLASTed alignment in the Output window.
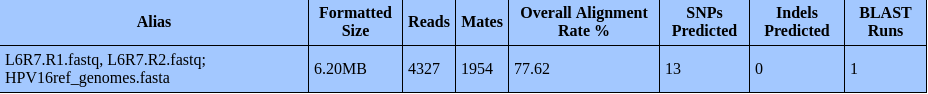
All the “BLAST Runs” for the given alignment, including their start and end positions, how many “suspicious” reads where actually submitted for BLASTing and the program used are viewable here.

Selecting a column will give a read by read breakdown.

For each read BLASTed, the position at which the read began aligning on the reference, the full read, BLAST’s top hit for the read, and the top hit’s E-value are given. Unaligned fragments in the read are displayed inside the read in red text at the position they actually appear in the read. Hovering over an unaligned fragment will show BLAST’s top hit for that fragment.
About BLASTing Alignments in PHAT
This functionality is effectively programmatic use of NCBI’s BLAST API. As such, we try to be good API citizens. BLAST requests are only submitted one at a time, and requests for status are submitted with appropriate delay based on BLAST’s API guidelines. The core API submission and polling logic is implemented here, based on BLAST’s sample Perl code.